Isobutanol Tolerance in S. cerevisiae
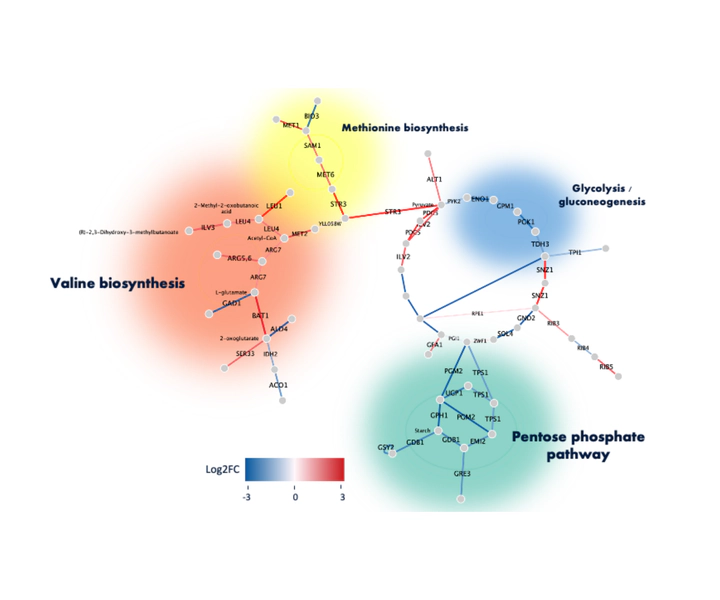 Top scoring active subnetwork in WT S. cerevisiae.
Top scoring active subnetwork in WT S. cerevisiae.Investigating pathways associated with isobutanol tolerance in S. cerevisiae to enhance biofuel production.
Summary
Background
The research focuses on identifying pathways associated with isobutanol tolerance in the yeast strain Saccharomyces cerevisiae. Isobutanol, a potential biofuel, is naturally produced by S. cerevisiae but in small amounts. The main challenge for industrial production is the yeast’s low tolerance to isobutanol accumulation, which hinders large-scale production. By understanding the pathways involved in isobutanol tolerance, the goal is to engineer yeast strains that can produce higher amounts of isobutanol.
Materials and Methods
The study utilised a transcriptomic dataset from a previous study by Kuroda et al. (2019), analysing differentially expressed (DE) genes in the S. cerevisiae GLN3∆ mutant strain under various conditions:
- WT (1.3% isobutanol) / WT (0%)
- GLN3∆ (0%) / WT (0%)
- GLN3∆ (1.3% isobutanol) / GLN3∆ (0%)
- GLN3∆ (1.3% isobutanol) / WT (1.3% isobutanol)
The data was processed using ShinyGATOM, a web tool for identifying regulated metabolic subnetworks, followed by functional annotation and pathway enrichment using KEGG and Reactome databases. The resulting subnetworks were visualised using Cytoscape to identify significant nodes and edges.
Results
- WT (1.3% isobutanol) / WT (0%): The top-scoring subnetwork contained pathways like valine and methionine biosynthesis (upregulated) and glycolysis/gluconeogenesis and pentose phosphate pathway (downregulated). The production of isobutanol was linked to valine breakdown.
- GLN3∆ (0%) / WT (0%): The subnetwork was too small for further analysis, indicating minimal changes in gene expression due to GLN3 deletion alone.
- GLN3∆ (1.3% isobutanol) / GLN3∆ (0%): The significant pathways were valine biosynthesis and starch and sucrose metabolism, with mixed up- and down-regulation.
- GLN3∆ (1.3% isobutanol) / WT (1.3% isobutanol): The largest subnetwork involved multiple pathways related to amino acid and secondary metabolite biosynthesis, and glycolysis/gluconeogenesis. The findings align with previous studies but noted the absence of some pathways (e.g., ion transport, cell wall biogenesis) due to limitations in the initial graph used.