Phenotype-Driven Variant Prioritisation Tools
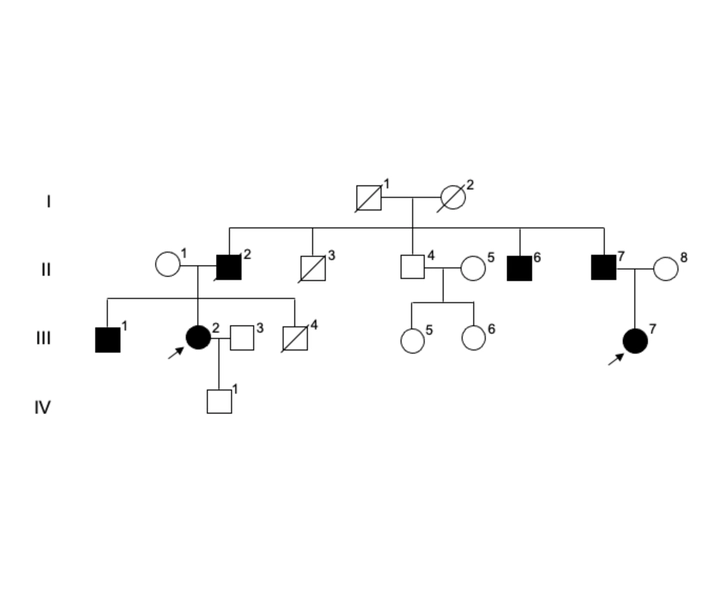 Familial pedigree of NG26.
Familial pedigree of NG26.Evaluation of computational tools for phenotype-driven variant prioritisation in Whole Exome Sequencing data.
Summary
Background
Mitochondrial disorders, particularly hereditary optic neuropathies (HON), have a complex genetic basis that is often difficult to decipher. These disorders can lead to optic atrophy and other neurological symptoms. The study addresses the challenge of identifying pathogenic mutations using high-throughput techniques like Whole Exome Sequencing (WES). However, the manual interpretation of Next Generation Sequencing (NGS) data is prone to errors, necessitating the use of computational algorithms for more accurate analysis.
Materials and Methods
The study analysed WES data from 29 individuals across 24 families, all exhibiting clinical signs of mitochondrial hereditary optic neuropathies. The cohort was screened at the IRCSS Institute of Neurological Sciences of Bologna. Various exome targets and phenotype-encoded terms (HPO) were used to annotate the patients’ phenotypes. The study evaluated four phenotype-driven variant prioritisation tools—The Exomiser, MutationDistiller, GenIO, and OVA—using patient data with known causative genes to assess their performance. The best-performing tools were then applied to identify potential pathogenic variants in genes linked to mitochondrial pathways or similar phenotypes.
Results
The study highlighted the utility of WES in routine clinical research for rare monogenic diseases, despite challenges like incomplete genomic coverage and the need to filter through a large number of variants. The variant prioritization tools tested helped streamline this process by predicting the pathogenicity of mutations based on various in silico methods. These tools allowed for the identification of candidate genes potentially responsible for the observed phenotypes in the cohort, facilitating more precise molecular diagnoses.