WASP: A pipeline for functional annotation based on AlphaFold structural models
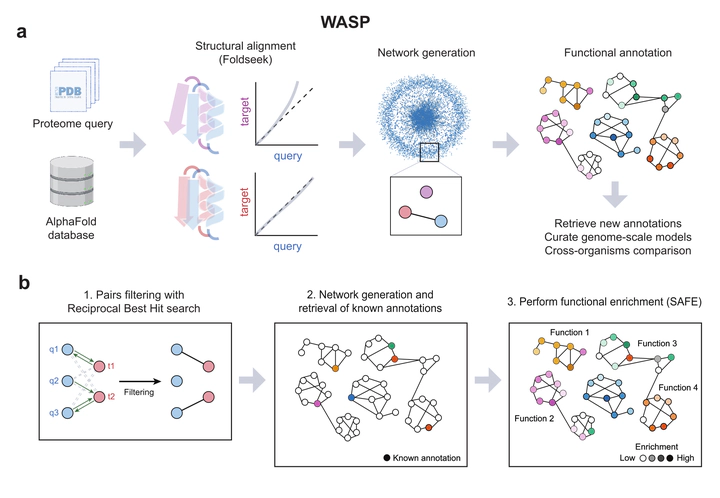 Overview of WASP workflow.
Overview of WASP workflow.Summary
WASP Overview: WASP is a novel pipeline for protein functional annotation starting from AlphaFold structure models. Using the output of state-of-the-art structural annotation tools, the proteome of a query organism is linked to structural homologs present in the AlphaFold database, creating a network whose topology is exploited to perform functional enrichment. In addition, WASP provides a holistic and comprehensive characterisation of proteins, using a diverse set of descriptors that provide a rich understanding of their molecular function.
Structural Homology Advantage: Previous methods primarily relied on sequence information to achieve annotation. However, protein structure is a major determinant in protein function, and it is known that structure is evolutionary more conserved while sequence divergence is more common. WASP is the first bioinformatic tool based on structure to annotate organisms at the whole-proteome level. WASP provides a user-friendly framework that allows advanced protein function prediction accessible to a broad range of researchers, from computational biologists to experimental scientists.
Benchmarking Performance: We benchmark WASP against multiple datasets and several state-of-the-art tools. We focus on classification based on EC numbers, a widespread system for enzyme activity categorisation. On hidden labels, WASP achieved superior F1 scores compared to state-of-the-art tools using sequence homology.
Improved Annotation and Industrial Application: WASP improved annotation in a cohort of industrially relevant organisms by 20-30% for previously uncharacterised proteins and successfully annotated a novel isolate of the yeast Yarrowia lipolytica. By improving annotation levels of high-potential organisms, WASP enhances the exploitation of their biological capabilities for various biotechnological applications.
Genome-Scale Metabolic Models (GEMs): WASP curated GEMs by gap-filling orphan reactions, identifying candidates for 75-100% of these reactions across 20 GEMs, based on plausible biological associations and providing insights into protein evolution. These results enhance our ability to perform quantitative biological analyses, enabling the construction of more accurate and predictive metabolic network models for better understanding and manipulation of biological systems.
Phylogenetic Tree Reconstruction: WASP provides superior clade discrimination in phylogenetic studies, offering precise mapping of evolutionary relationships and functional diversities among proteins, and deeper insights into protein evolution within and across species.